Balbharti Maharashtra State Board 12th Biology Important Questions Chapter 4 Molecular Basis of Inheritance Important Questions and Answers.
Maharashtra State Board 12th Biology Important Questions Chapter 4 Molecular Basis of Inheritance
Multiple Choice Questions
Question 1.
How many of the following characteristics are shown by the R-strain of Streptococcus pneumonia? Avirulent, Smooth, Pathogenic, Capsulated ………………..
(a) One
(b) TWo
(c) Three
(d) Four
Answer:
(a) One
Question 2.
Griffith obtained …………….. from the blood of the dead mice.
(a) dead S-strain bacteria
(b) live R-strain bacteria
(c) dead R-strain bacteria
(d) live S-strain bacteria
Answer:
(d) live S-strain bacteria
![]()
Question 3.
Oswald T. Avery, Colin M. MacLeod and Maclyn McCarty demonstrated that ………………..
(a) transformation of live S-strain bacteria into R-strain type was because of DNA of bacteria of S-strain.
(b) the transforming substance was either a protein or RNA.
(c) only DNA was able to transform harmless R-strain into virulent S-strain.
(d) when DNA isolated from S-strain bacteria, was digested with DNase, the transformation occurred.
Answer:
(c) only DNA was able to transform harmless R-strain into virulent S-strain
Question 4.
Which of the following was NOT observed in Hershey and Chase experiment?
(a) Viruses grown in the presence of radioactive sulphur, had radioactive protein but not radioactive DNA.
(b) Radioactive ‘P’ remained in suspension.
(c) Only radioactive ‘P’ was found inside the bacterial cells in the pellet.
(d) Viruses grown in the presence of radioactive phosphorus contained radioactive DNA but not radioactive proteins.
Answer:
(b) Radioactive ‘P’ remained in suspension.
Question 5.
Enzymes like ……………….. and DNA topoisomerase-I, play important role in maintaining super-coiled state in prokaryotic DNA.
(a) DNA ligase
(b) DNA gyrase
(c) RNA polymerase
(d) None of these
Answer:
(b) DNA gyrase
Question 6.
Histone octamer of nucleosome has two molecules, each of ……………….. proteins.
(a) H2A, H2B, H3 and H4
(b) H2A, H2B, H3 and H1
(c) H2A, H2B. H3A and H3B
(d) H1A, H2B, H3A and H4
Answer:
(a) H2A, H2B, H3 and H4
Question 7.
Select the CORRECT statement.
(a) Euchromatin is mainly located near centromere and telomeres.
(b) Heterochromatin replicates at faster rate than euchromatin.
(c) Heterochromatin has 2 to 3 times more DNA than in the euchromatin.
(d) Heterochromatin is lightly stained region of chromonema.
Answer:
(c) Heterochromatin has 2 to 3 times more DNA than in the euchromatin
Question 8.
A DNA molecule in which both strands have 14N is allowed to replicate in an environment containing 15N. What will be the exact number of DNA molecules that contain the 14N after three replications?
(a) One
(b) Two
(c) Four
(d) Eight
Answer:
(b) Two
Question 9.
As the base sequence present on one strand of DNA decides the base sequence of other 8 strand, this strand is considered as ………………..
(a) descending strand
(b) leading strand
(c) lagging strand
(d) complementary strand
Answer:
(d) complementary strand
Question 10.
In prokaryotes ……………….. recognizes the promoter sequence.
(a) alpha factor
(b) rho factor
(c) theta factor
(d) sigma factor
Answer:
(d) sigma factor
Question 11.
If the base sequence in DNA is 5′ AAAA 3′, then the base sequence in m-RNA is ………………..
(a) 5′ UUUU 3′
(b) 3′ UUUU 5′
(c) 5′ AAAA 3′
(d) 3′ TTTT 5′
Answer:
(c) 5′ AAAA 3′
Question 12.
During capping, methylated guanosine tri¬phosphate is added to 5′ end of ………………..
(a) m-RNA
(b) t-RNA
(c) hnRNA
(d) r-RNA
Answer:
(c) hnRNA
Question 13.
If each codon has two nucleotides, then there will be ……………….. codons, which can encode for only …………….. different types of amino acids.
(a) 16, 16
(b) 16, 20
(c) 20, 16
(d) 64, 64
Answer:
(a) 16, 16
Question 14.
What would happen if in a gene encoding a polypeptide of 50 amino acids, 25th codon (UAU) is mutated to UAA?
(a) A polypeptide of 24 amino acids is formed.
(b) Two polypeptides of 24 and 25 amino acids will be formed.
(c) A polypeptide of 49 amino acids is formed.
(d) A polypeptide of 25 amino acids is formed.
Answer:
(a) A polypeptide of 24 amino acids is formed.
Question 15.
A strand of DNA has following base sequence – 3′ AAAAGTGAATAGTGA 5′. On transcription it produces an m-RNA. Which of the following anticodon of t-RNA recognizes the third codon of this m-RNA?
(a) AAA
(b) CUG
(c) AAG
(d) CTG
Answer:
(c) AAG
Question 16.
Polynucleotide chain consisting of only CUA repeats will give polypeptide chain with only one amino acid ………………..
(a) tryptophan
(b) leucine
(c) serine
(d) methionine
Answer:
(b) leucine
Question 17.
Select the INCORRECT statement.
(a) Dr. Khorana prepared polyribo-nucleotides chains with known repeated sequences of two or three nucleotides by using synthetic DNA.
(b) M. Nirenberg and Matthaei synthesized artificial m-RNA which was a homopolymer of uracil ribonucleotides.
(c) Enzyme polynucleotide phosphorylase polymerizes RNA with defined sequences in a template-dependent manner.
(d) Evidence for triplet nature of geneticcode, was given by Crick (1961) using “frame-shift mutation”.
Answer:
(c) Enzyme polynucleotide phosphorylase polymerizes RNA with defined sequences in a template-dependent manner.
Question 18.
…………… is/are based on complementarity principle.
(a) Replication and translation
(b) Replication and transcription
(c) Translation
(d) Only replication
Answer:
(b) Replication and transcription
Question 19.
Cysteine has codons, while isoleucin has ……………….. codons.
(a) two, three
(b) three, two
(c) two, four
(d) four, two
Answer:
(a) two, three
Question 20.
Out of 64 codons, only 61 code for the 20 different amino acids. This is known as ……………….. of genetic code.
(a) non-ambiguity
(b) overlapping nature
(c) ambiguity
(d) degeneracy
Answer:
(d) degeneracy
Question 21.
Mutation that results in Sickle-cell anaemia is a ………………..
(a) deletion
(b) frame-shiftmutation
(c) point mutation
(d) insertion
Answer:
(c) point mutation
Question 22.
Initiator charged t-RNA occupies the ……………….. of ribosome first.
(a) A-site
(b) P-site
(c) E-site
(d) either A-site or P-site
Answer:
(b) P-site
Question 23.
It takes ……………….. for formation of peptide bond.
(a) 10 seconds
(b) 0.1 second
(c) less than 0.1 second
(d) 60 seconds
Answer:
(c) less than 0.1 second
Question 24.
Anticodon and codon bind by ………………..
(a) glycosidic bond
(b) hydrogen bond
(c) phosphodiester bond
(d) none of these
Answer:
(b) hydrogen bond
Question 25.
The UTRs are present at ………………..
(a) 5′-end, before start codon and at 3′-end, after stop codon of m-RNA
(b) 5′-end, before start codon and at 3′-end, after stop codon of t-RNA
(c) only at 3′-end, after stop codon of m -RNA
(d) only at 5′-end, before start codon of m-RNA
Answer:
(a) 5′-end, before start codon and at 3′-end, after stop codon of m-RNA
Question 26.
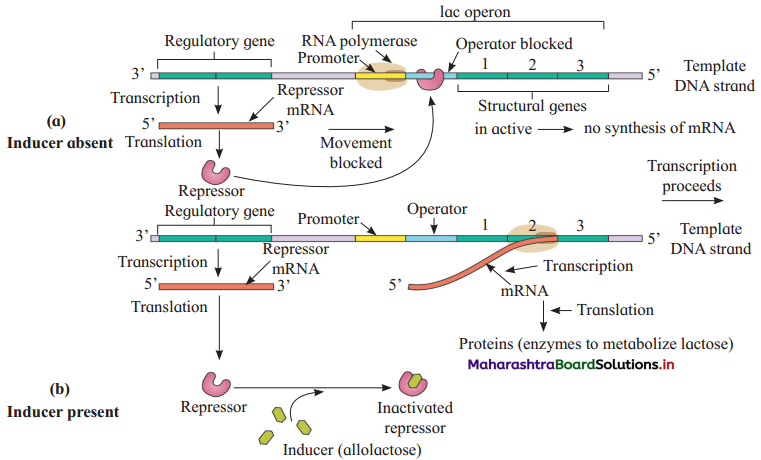
The action of structural genes is regulated by …………….. site with the help of a …………….. protein.
(a) operator, inducer
(b) operator, repressor
(c) regulator, repressor
(d) regulator, inducer
Answer:
(b) operator, repressor
Question 27.
Repressor protein is produced by the action of ………………..
(a) gene z
(b) gene y
(c) gene i
(d) gene o
Answer:
(c) gene i
Question 28.
Select the correct pair.
(a) Gene z – Transacetylase
(b) Gene y – Beta-galactocidase
(c) Gene a – Beta-galactoside permease
(d) Gene I – Repressor
Answer:
(d) gene I – repressor
![]()
Question 29.
Structural genomics involves ……………….. of genome.
(a) mapping
(b) sequencing
(c) analysis
(d) all of these
Answer:
(d) all of these
Question 30.
The technique of transferring DNA fragments separated on agarose gel to a synthetic nitrocellulose membrane is known as ………………..
(a) Southern blotting
(b) Autoradiography
(c) Southern hybridization
(d) None of these
Answer:
(a) Southern blotting
Question 31.
Sequence of various steps in DNA fingerprinting is ………………..
i. Southern blotting.
ii. Restriction digestion
iii. Agarose gel electrophoresis
iv. DNA isolation
v. Photography.
vi. Selection of DNA probe
vii. Hybridization
(a) iv, iii, ii, i, v, vi, vii
(b) iv. v, iii, i, vi, vii, ii
(c) iv, ii, iii, i, vi, vii, v
(d) ii, iii, iv, i, vi, vii,
Answer:
(c) iv, ii, iii, i, vi. vii, v
Match the Columns
Question 1.
| Column A | Column B |
| (1) Frederick Griffith | (a) Test tube assay |
| (2) Avery, McCarty and MacLeod | (b) Streptococcus pneumoniae |
| (3) Alfred Hershey and Martha Chase | (c) E. coli |
| (4) Meselson and Stahl | (d) Bacteriophages |
Answer:
| Column A | Column B |
| (1) Frederick Griffith | (b) Streptococcus pneumoniae |
| (2) Avery, McCarty and MacLeod | (a) Test tube assay |
| (3) Alfred Hershey and Martha Chase | (d) Bacteriophages |
| (4) Meselson and Stahl | (c) E. coli |
Classify the following to form Column B as per the category given in Column A
Question 1.
(i) UUU
(ii) CUA
(iii) UAA
(iv) AUG
(v) UAG
(vi) UGA
| Column A | Column B |
| 1. Initiator codon | ————– |
| 2. Stop codons | ————- |
| 3. Codon that codes for Phenyl alanin | ————– |
| 4. Codon that codes for leucine | ————- |
Answer:
| Column A | Column B |
| 1. Initiator codon | (iv) AUG |
| 2. Stop codons | (iii) UAA, (v) UAG, (vi) UGA |
| 3. Codon that codes for Phenyl alanin | (i) UUU |
| 4. Codon that codes for leucine | (ii) CUA |
Question 2.
(i) Severo Ochoa
(ii) F. Jacob and J. Monod
(iii) Temin and Baltimore
(iv) H. Winkler
(v) T. H. Roderick
(vi) R Kornberg
| Column A | Column B |
| (1) Lac Operon | ————– |
| (2) Central dogma in retroviruses | ————- |
| (3) Coined the term Genome | ————– |
| (4) Coined the term Genomics | ————- |
| (5) Enzymatic synthesis of RNA | ————- |
| (6) DNA is associated with histones and non-histones | ————– |
Answer:
| Column A | Column B |
| (1) Lac Operon | (ii) E Jacob and J. Monod |
| (2) Central dogma in retroviruses | (iii) Temin and Baltimore |
| (3) Coined the term Genome | (iv) H. Winkler |
| (4) Coined the term Genomics | (v) T. H. Roderick |
| (5) Enzymatic synthesis of RNA | (i) Severo Ochoa |
| (6) DNA is associated with histones and non-histones | (vi) R. Kornberg |
Very Short Answer Questions
Question 1.
What are the two types of bacteria used by F. Griffith and which one out of these is avirulent?
Answer:
S-type and R-type strains of Streptococcus penumoniae were used by F. Griffith and out of these R-type is avirulent.
Question 2.
Enlist the characteristics of S-strain pneumoniae.
Answer:
S-strain pneumoniae are virulent, smooth and encapsulated.
Question 3.
What is the bacteriophage?
Answer:
Bacteriophage is a virus that infects bacterium and injects its genetic material in the bacterium.
Question 4.
What is the length of DNA double helix molecule in a typical mammalian cell?
Answer:
The length of DNA double helix molecule in a typical mammalian cell is approximately 2.2 meters.
Question 5.
What is the approximate size of a typical nucleus ?
Answer:
Approximate size of a typical nucleus is 10-6 m.
Question 6.
What is size of E. coli cell?
Answer:
The size of E. coli cell size is 2-3 µm.
Question 7.
What determines the charge on protein molecules?
Answer:
A protein acquires its charge depending upon the abundance of amino acid residues with charged side chains.
Question 8.
What is nucleosome core?
Answer:
Nucleosome core is a histone octamer.
Question 9.
Where is H1 histone present?
Answer:
H1 histone binds the DNA thread where it enters and leaves the nucleosome.
Question 10.
How are solenoid fibres formed?
Answer:
Six nucleosomes get coiled and then form solenoid that looks like coiled telephone wire of 30 nm diameter (300Å).
Question 11.
How is chromatin fibre formed?
Answer:
Supercoiling of solenoid fibre forms a looped structure called chromatin fibre.
Question 12.
What is NHC?
Answer:
NHC stands for Nonhistone Chromosomal proteins.
Question 13.
List as many different enzyme activities required during DNA synthesis as you can.
Answer:
Phosphorylase, Helicase, DNA polymerase, Primase, DNA ligase, Super helix relaxing enzyme, Topoisomerase (gyrase) are different enzymes required during DNA synthesis.
Question 14.
How many replicons are present in prokaryotes and eukaryotes respectively?
Answer:
Prokaryotes have one replicon. Several replicons in tandem are present in eukaryotes.
Question 15.
What is the function of SSBP?
Answer:
During replication of DNA SSBP proteins remain attached to both the separated strands and prevent them from coiling back.
Question 16.
During which phases of cell cycle, transcription occurs in the nucleus?
Answer:
Transcription occurs in the nucleus during G1 and G2 phases of cell cycle.
Question 17.
Which strand of transcription unit gets transcribed ?
Answer:
DNA strand having 3’ → 5’ polarity acts as template strand and it gets transcribed.
Question 18.
What is a cryptogram?
Answer:
Cryptogram is a genetic code consisting of triplet codons on m-RNA that code for a specific amino acids.
Question 19.
What is meant by the polarity of genetic code?
Answer:
Genetic code is always read in 5′ → 3’ direction. This is called polarity of genetic code.
Question 20.
Which mutation can result in changes in the reading frame?
Answer:
Insertion or deletion of one or two bases changes the reading frame from the point of insertion or deletion.
Question 21.
Which mutation can result in insertion or deletion of amino acids, but reading frame remains unaltered?
Answer:
Insertion or deletion of three or multiples of three bases results in insertion or deletion of amino acids and reading frame remains unaltered from that point onwards.
Question 22.
Which molecule serves as an intermediate molecules between DNA and protein during proteins synthesis?
Answer:
RNA serves as an intermediate molecule between DNA and protein during proteins synthesis.
![]()
Question 23.
What is the function of a groove present between two subunits of ribosome in eukaryotes ?
Answer:
The groove present between two subunits of ribosomes in eukaryotes protects the polypeptide chain from the action of cellular enzymes and also protects m-RNA from the action of nucleases.
Question 24.
Enlist different steps of protein synthesis.
Answer:
Steps in protein synthesis are:
- Transcription
- Activation of amino acids and formation of charged t-RNAs,
- Synthesis of polypeptide chain:
- initiation
- elongation and
- termination of polypeptide chain.
Question 25.
What is translocation?
Answer:
During elongation of polypeptide chain, the ribosome moves along the m-RNA in stepwise manner from start codon to stop codon (5′ → 3′), 1 codon ahead each time t, his movement is called translocation and due to this t-RNA carrying a dipeptide at A-site of the ribosome moves to the p-site.
Question 26.
What is meant by inducible enzymes?
Answer:
Bacteria like E.coli adapt to their chemical environment by synthesizing certain enzymes depending upon the substrate present. Such adaptive enzyme is called inducible enzyme.
Question 27.
What is meant by induction and inducer?
Answer:
A set of genes are switched on when a new substrate is to be metabolized. This phenomenon is called induction and small molecule responsible for this is known as inducer.
Question 28.
What is the role of a repressor gene?
Answer:
The role of a repressor gene is to produce repressor protein. Repressor binds with operator gene and this prevents transcription of structural genes in the operon.
Question 29.
Which molecule does act as inducer molecule in lac operon?
Answer:
Allolactose acts as inducer molecule in lac operon.
Question 30.
In which condition, lac operon is switched off?
Answer:
If E.coli bacteria do not have lactose in the surrounding medium as a source of energy, lac operon is switched off.
Question 31.
What lac operon consists of?
Answer:
Lac operon consists of a regulator promoter, operator and three structural genes z, y and a.
Question 32.
Which gene acts as a regulatory gene in lac operon?
Answer:
Repressor protein is produced by the action of gene i (inhibitor). This gene acts as a regulator gene.
Question 33.
When was Human Genome Project started ? When was it completed ?
Answer:
The Human Genome Project was started in 1990 and was completed in 2003.
Question 34.
What is functional genomics?
Answer:
Functional genomics is a branch of genomics that involves the study of functions of all gene sequences and their expressions in organisms.
Question 35.
What is the advantage of sequencing of genomes of non-human organisms?
Answer:
Sequencing of genomes of non-human model organisms allows researchers to study gene functions in these organisms. Since human beings possess many genes which are like those of flies, roundworms and mice, comparative studies will lead to greater understanding of human evolution.
Question 36.
What are VNTRs?
Answer:
Variable Number of Tandem Repeats (VNTRs) are unusual sequences of 20-100 base pairs, which are repeated several times and are arranged tandency.
Question 37.
Do different organisms have the same DNA?
Answer:
Different organisms differ in their DNA sequence.
Question 38.
What is the amino acid sequence encoded by base sequence UCA, UUU, UCC, GGG, AGU of an m-RNA segment?
Answer:
The amino acid sequence: Ser-Phe – Ser – Gly- Ser
Give definitions of the following
Question 1.
Replicon
Answer:
The unit of DNA in which replication occurs is known as replicon.
Question 2.
Transcription
Answer:
Transcription is defined as the process of copying of genetic information from template strand of DNA into a complementary single stranded RNA transcript.
Question 3.
Gene
Answer:
Gene is defined as the DNA sequence coding for m-RNA/ t-RNA or r-RNA.
Question 4.
Cistron
Answer:
Cistron is defined as a segment of DNA coding for a polypeptide.
Question 5.
Monocistronic gene
Answer:
Gene is called monocistronic, when there is a single structural gene in one transcription unit.
Question 6.
Polycistronic gene
Answer:
Gene is called polycistronic, when there is a set of various structural genes in one transcription unit.
Question 7.
Interrupted or Split genes
Answer:
Interrupted or split genes are the structural genes in eukaryotes which have both exons and introns.
Question 8.
Exons
Answer:
Exons are the coding sequences or express sequences in DNA/hnRNA/ m-RNA.
Question 9.
Introns
Answer:
Introns are the non-coding sequences in DNA or hnRNA.
Question 10.
Anticodon
Answer:
Anticodon is a triplet of nucleotides present on the anticodon loop of t-RNA, which is complementary to codon on m-RNA.
Question 11.
Mutation
Answer:
Mutation is a sudden heritable change in the DNA sequence that results in the change of genotype.
Question 12.
Translation
Answer:
Translation is the process in which sequence of codons of m-RNA is decoded and accordingly amino acids are added in specific sequence to form a polypeptide on ribosomes.
Question 13.
Genomics
Genomics is the study of genomes through analysis, sequencing and mapping of genes along with the study of their functions.
Question 14.
Repressors
Answer:
Repressors are proteins which are able to bind the operator region of operon and prevent the RNA polymerase from transcribing the operon.
Name the following
Question 1.
Enzyme that cleaves DNA.
Answer:
DNase
Question 2.
Enzyme that cleaves proteins.
Answer:
Protease
Question 3.
Enzyme involved in activation of nucleotides.
Answer:
Phosphorylase
Question 4.
Enzyme involved in unwinding of DNA.
Answer:
Helicase
Question 5.
Enzyme involved in synthesis of DNA.
Answer:
DNA polymerase
Question 6.
Enzyme involved in joining of Okazaki fragments.
Answer:
DNA ligase
Question 7.
Enzyme involved in synthesis of RNA primer.
Answer:
Primase
Question 8.
Enzyme involved in removal of RNA primer.
Answer:
DNA polymerase
Question 9.
Enzyme involved in replacement of gaps in prokaryotes.
Answer:
DNA polymerase – I
![]()
Question 10.
Enzyme involved in replacement of gaps in eukaryotes.
Answer:
DNA polymerase α
Question 11.
Enzyme involved in formation of double helix in daughter DNA molecules.
Answer:
Topoisomerase
Question 12.
Enzyme involved in releasing strain created by unwinding of DNA.
Answer:
Super helix relaxing enzyme
Question 13.
Enzyme involved in synthesis of hnRNA, m-RNA.
Answer:
RNA polymerase – II
Question 14.
Enzyme involved in synthesis of t-RNA, snRNA.
Answer:
RNA polymerase – III
Question 15.
Enzyme involved in synthesis of r-RNA
Answer:
RNA polymerase – I
Question 16.
Enzyme involved in polymerizing RNA in template independent manner.
Answer:
Polynucleotide phosphorylase
Question 17.
Enzyme involved in peptide bond synthesis.
Answer:
Ribozyme
Question 18.
Enzyme involved in Cutting DNA at specific sites.
Answer:
Restriction endonuclease
Question 19.
Name the initiator codon of protein synthesis.
Answer:
AUG is the initiator codon of protein synthesis.
Question 20.
Name three binding sites of ribosome.
Answer:
Three binding sites for t-RNA on ribosomes are P-site (peptidy t-RNA-site), A-site (aminoacyl – t-RNA-site) and E-site (exit site).
Question 21.
Name the different structural genes in sequence of lac operon.
Answer:
There are 3 structural genes in the sequence lac-Z, lac-Y and lac-A.
Question 22.
Name the organisms whose genomes have been sequenced?
Answer:
The genomes of several organisms such as bacteria e.g. E.coli, Caenorhabditis elegans (a free living non-pathogenic nematode), Saccharomyces cerevisiae (yeast), Drosophila (fruit fly), plants (rice and Arabidopsis), Mus musculus (mouse), etc. have been sequenced.
Give Significance/Functions of the following
Question 1.
DNA.
Answer:
- DNA regulates and controls all the cellular activities.
- It replicates and gets distributed equally to the daughter cells when the cell divides.
- It is a carrier of genetic information.
- Heterocatalytic function : DNA directs the synthesis of chemical molecules other than itself. E.g. Synthesis of RNA (transcription), synthesis of protein (Translation), etc.
- Autocatalytic function : DNA directs the synthesis of DNA itself. E.g. Replication.
- DNA is a master molecule of a cell that initiates, guides, regulates and controls the process of protein synthesis.
Question 2.
Proteins.
Answer:
Proteins serve as structural components, enzymes and hormones.
Distinguish between the following.
Question 1.
Euchromatin and Heterochromatin.
Answer:
| Euchromatin | Heterochromatin. |
| 1. Euchromatin is loosely packed region of the chromatain. | 1. Heterochromatin is densely packed region of the chromatin. |
| 2. Euchromatin stains lightly. | 2. Heterochromatin stains darkly. |
| 3. Euchromatin is transcriptionally active region of the chromatin. | 3. Heterochromatin is transcriptionally inactive region of the chromatin. |
Question 2.
DNA in prokaryotic and eukaryotic cells.
Answer:
| DNA in prokaryotes | DNA in eukaryotic |
| 1. It is present in the cytoplasm. | 1. It is present in the nucleus. |
| 2. It is not associated with histones. | 2. It is associated with histones. |
| 3. It is circular. | 3. It is linear. |
| 4. Genes do not contain introns. | 4. Genes contain introns along with exons. |
| 5. Genes are polycostronic. | 5. Genes are monocistronic. |
Question 3.
DNA and RNA
Answer:
| DNA | RNA |
| 1. DNA is deoxyribonucleic acid. | 1. RNA is ribonucleic acid. |
| 2. DNA is double stranded, helical molecule. | 2. RNA is single stranded molecule. |
| 3. In DNA, there is deoxyribose sugar. | 3. In RNA, there is ribose sugar. |
| 4. The pyrimidine nitrogen bases are cytosine and thymine. | 4. The pyrimidine nitrogen bases are cytosine and uracil. |
| 5. DNA is the genetic material in all types of organisms. | 5. RNA is genetic material in few viruses only. |
| 6. In eukaryotic cells, DNA is present in nucleus. | 6. In eukaryotic cells, RNA is present in nucleus as well as cytoplasm. |
| 7. The number of purine : pyrimidine ratio is always 1 : 1 in DNA molecule. | 7. The number of purine : pyrimidine ratio may not be 1 : 1 in RNA molecule. |
| 8. DNA sends the codon for the synthesis of proteins, but otherwise it does not participate in the protein synthesis. | 8. RNA takes part in the protein synthesis through transcription and translation. |
Question 4.
m-RNA, t-RNA and r-RNA.
Answer:
| m-RNA | t-RNA | r-RNA. |
| 1. m-RNA is a simple molecule which shows linear structure without any folds. | 1. t-RNA is a single stranded molecule. There is regular pattern of folding shown by this molecule. | 1. r-RNA is a single stranded RNA that is variously folded upon itself. In the folded regions it shows complementary base pairing. |
| 2. It performs the function of transcription during protein synthesis. | 2. It performs the function of transferring the amino acids during translation. | 2. This RNA remains associated with the ribosomes permanently. It gives the binding site for m-RNA during the process of protein synthesis. It also orients the m-RNA molecule so as to read the message on codons properly. |
| 3. Of the total cellular RNA, m-RNA forms 3-5%. | 3. Of the total cellular RNA, t-RNA forms about 10-20%. | 3. Of the total cellular RNA, r-RNA forms 80% |
| 4. It has molecular weight of about 5,00,000. | 4. t-RNA is the smallest RNA having only 73-93 nucleotides and has molecular weight of about 23,000 to 30,000 daltons. | 4. Molecular weight is about 40,000 to 100,000 daltons. |
Give Reasons
Question 1.
Nuclein was called as nucleic acid.
Answer:
Nuclein had acidic properties and it was isolated from nucleus. Hence, it was called as nucleic acid.
Question 2.
Initially proteins (and not DNA) were considered as genetic material.
Answer:
- Proteins are large, complex molecules and store information required to govern cell metabolism. Hence it was assumed that variations found in species were caused by proteins.
- On the other hand, DNA was considered as a small, simple molecule whose composition does not vary much among species.
- Variations in the DNA molecules are different than the variation in shape, electrical charge and function shown by proteins.
- Hence, initially proteins (and not DNA) were considered as genetic material.
Question 3.
On injecting a mixture of heat-killed S-bacteria and live R bacteria, the mice died.
Answer:
- Griffith obtained live S-strain bacteria from the blood of the dead mice.
- In a mixture of live R-bacteria and heat killed S-bacteria, live R-strain bacteria picked up something (transforming principle) from the heat-killed S bacterium and got changed into S-type.
- Transforming principle allowed R-type bacteria to synthesize capsule and thus they became virulent.
- Hence, on injecting a mixture of heat-killed S bacteria and live R bacteria, the mice died.
Question 4.
Viruses obtained by infecting bacteria having radioactive phosphorus contained radioactive DNA (labelled DNA), but not radioactive proteins.
Answer:
Viruses obtained by infecting bacteria having radioactive phosphorus, contained radioactive DNA (labelled DNA). but not radioactive proteins because DNA contains phosphorus (labelled DNA) but proteins do not.
Question 5.
Viruses obtained by infecting bacteria having radioactive sulphur contained radioactive protein but not radioactive DNA.
Answer:
Viruses obtained by infecting bacteria having radioactive sulphur contained radioactive protein but not radioactive DNA because DNA does not contain sulphur and proteins contain sulphur.
Question 6.
In bacteria, m-RNA does not require any processing.
Answer:
In bacteria, m-RNA does not require any processing because it has no introns and it is synthesized in cytoplasm.
Question 7.
Eukaryotic DNA is condensed and supercoiled.
Answer:
- In a typical mammalian cell, length of DNA double helix is approximately 2.2 metres.
- The size of typical nucleus is approximately 10-6 m
- Such a long DNA molecule has to be fitted in small nuclear space.
- Therefore, DNA is highly condensed, coiled and supercoiled so that it can be accommodated in the nucleus.
![]()
Question 8.
During translation, complementarity principle is not applicable.
Answer:
During translation, complementarity principle is not applicable as, genetic information is transferred from a polymer of nucleotides to a polymer of amino acids.
Question 9.
Protein synthesis is the most important and essential activity in the living cells.
Answer:
- Proteins play a significant role in the metabolism of living cells.
- The actual phenotypic expression of living cells is dependent on the biochemical reactions.
- Each biochemical reaction needs a specific enzyme for its initiation and completion. All the enzymes are proteins.
- In a cell there are many structural proteins too. Thousands of structural and catalytic proteins are constantly required within the cell at all times.
- Many functional proteins like hormones are also important for metabolism. Thus, for the synthesis of all such proteins, protein synthesis has become the most important and essential activity of the living cell.
Question 10.
Only 20 amino acids are considered as standard.
Answer:
- It was believed that there are total 20 amino acids in the living world. But 21st amino acid called selenocysteine was discovered later.
- This amino acid is coded by UGA which is usually a termination codon.
- In both prokaryotic and eukaryotic cells polypeptide chains contain 100-300 amino acids and they are formed by specific arrangement of 21 amino acids.
- But formation of selenocysteine requires the availability of element selenium in the cells.
- Therefore, only 20 amino acids are considered as standard.
Write Short Notes on the following
Question 1.
Friedrich Miescher’s nuclein
Answer:
- Nuclein is an acidic substance, having high phosphorus content and it was isolated by Friedrich Miescher in 1869, from the nuclei of pus cells.
- As Nuclein had acidic properties and it was isolated from nucleus, it was called as nucleic acid.
- E Miescher started working with white blood cells (the major component of pus). He used a salt solution to wash the pus off the bandages. He lysed the cells by adding a weak alkaline solution and isolated nucleic acid from nuclei that precipitated out of the solution.
- By the early 1900s, it was known that Miescher’s nuclein was a mixture of proteins and nucleic acids (DNA and RNA).
Question 2.
Packaging in Prokaryotes
Answer:
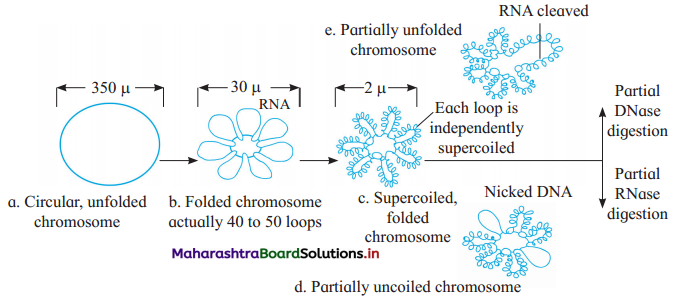
- Size of cell in E. coli size is 2-3µ.
- The nucleoid is small, circular, highly folded, naked DNA (1100 µm long in perimeter and contains about 4.6 million base pairs).
- When the negatively charged DNA becomes circular, the size reduces to 350 µm in diameter.
- Folding/looping (40-50 domains (loops) further reduce it to 30 µm in diameter.
- RNA connectors assist in loop formation.
- Further coiling and super coiling of each domain reduces the size to 2 µm in diameter.
- This coiling (packaging) is assisted by positively charged HU (Histone like DNA binding proteins) proteins and enzymes like DNA gyrase and DNA topoisomerase-I, which maintain supercoiled state.
Question 3.
Experimental confirmation of semiconservative replication of DNA.
Answer:
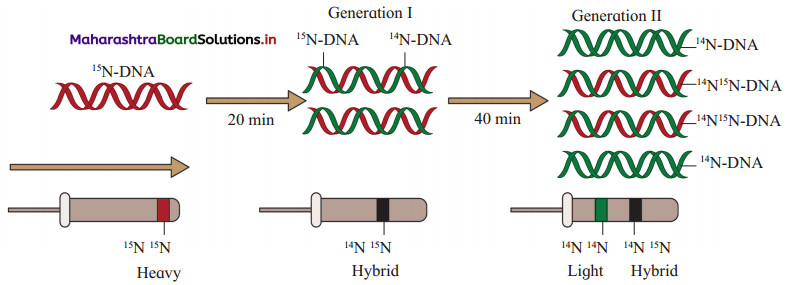
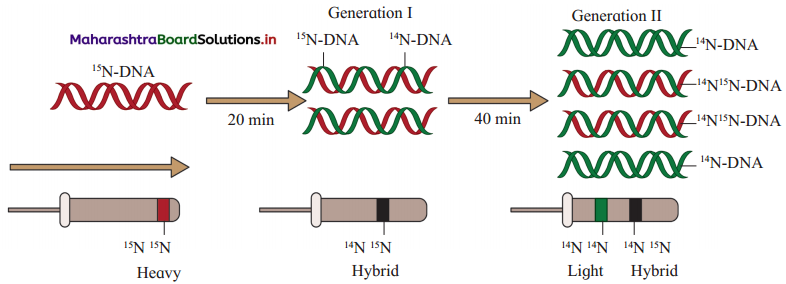
(1) Matthew Meselson and Franklin Stahl (1958) used equilibrium – density – gradient – centrifugation technique to experimentally prove semiconservative DNA replication.
(2) They cultured bacteria E.coli in the medium containing 14N (light nitrogen). They obtained equilibrium density gradient band by using 6M CsCl2. The position of this band is recorded.
(3) E. coli cells were then transferred to 15N medium (heavy isotopic nitrogen) and allowed to replicate for several generations. At equilibrium point density gradient band was obtained, by using 6M CsCl2. The position of this band is recorded.
(4) The heavy DNA (15N) molecule can be distinguished from normal DNA by centrifugation in a 6M Cesium chloride (CsCl2) density gradient. At the equilibrium point 15N DNA will form a band. In this both the strands of DNA are labelled with 15N.
(5) Such E. coli cells were then transferred to another medium containing 14N i.e. normal (light) nitrogen. After first generation, the density gradient band for 14N 15N was obtained and its position was recorded. After second generation, two density gradient bands were obtained – one at 14N 15N position and other at 14N position.
(6) The position of bands after two generations clearly proved that DNA replication is semiconservative.
Question 4.
Central Dogma of molecular biology.
Answer:
(1) Central dogma of molecular biology was postulated by EH.C. Crick in 1958.
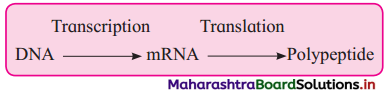
(2) DNA gets transcribe to form m-RNA, m-RNA acts as a messenger and gets translated to form a polypeptide chain (protein) having specific amino acid sequence.
(3) This unidirectional flow of information from DNA to RNA and from RNA to proteins is referred as central dogma of molecular biology.
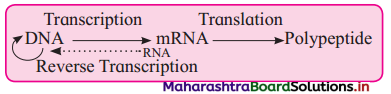
(4) Temin (1970) and Baltimore (1970) : Central dogma in retroviruses.
Question 5.
Processing of hnRNA.
Answer:
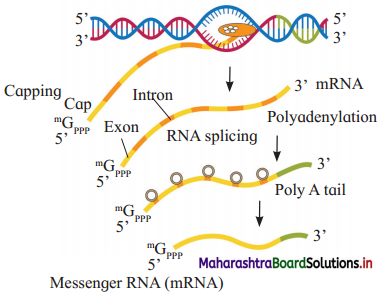
- Eukaryotes have split genes.
- Primary transcript or hnRNA is non-functional and contains both exons and introns.
- Processing of hnRNA results in functional m-RNA.
- The fully processed hnRNA is called m-RNA.
- m-RNA comes out of the nucleus through nuclear pore for getting translated.
hnRNA undergoes capping, tailing and splicing.
- Capping : Methylated guanosine tri¬phosphate is added to 5’ end of hnRNA.
- Tailing : Polyadenylation take place at 3′ end.
- Splicing : It is removal of introns.
- DNA ligase joins exons in a definite sequence (order).
Question 6.
Cracking of genetic code.
Answer:
- M. Nirenberg and Matthaei synthesized artificial poly-U m-RNA.
- Using this synthetic poly-U m-RNA and cell free system of protein synthesis, a small polypeptide consisting of only amino acid phenylalanine was obtained.
- It suggested that UUU codes for phenylalanine.
- Dr. Har Gobind Khorana devised a technique for synthesis of artificial m-RNA with repeated sequences of known nucleotides.
- He synthesized artificial RNA consisting of known repeated sequences of two or three nucleotides. E.g. CUC, UCU, CUC, UCU, by using synthetic DNA.
- This resulted in formation of polypeptide chain consisting of alternate amino acids leucine and serine.
- Synthetic RNA consisting of CUA, CUA, CUA, CUA repeats gave polypeptide chain with only one amino acid – leucine.
- Severo Ochoa established that the enzyme (polynucleotide phosphorylase) also helps in polymerizing RNA of defined sequences in a template-independent manner (i.e. enzymatic synthesis of RNA).
- Thus all the 64 codons in the dictionary of genetic code were deciphered.
Question 7.
Types of mutations.
Answer:
- Chromosomal mutations : Loss (deletion) or gain (insertion/duplication) of a segment of DNA results in alteration in the chromosome.
- Point mutations : They involve change in a single base pair of DNA. E.g. Mutation that results in Sickle-cell anaemia.
- Deletion or insertion of base pairs of DNA : It causes frame-shift mutations or deletion mutation.
- Insertion or deletion of one or two bases changes the reading frame from the point of insertion or deletion.
- Insertion or deletion of three or multiples of three bases (insert or delete) results in insertion or deletion of amino acids and reading frame remains unaltered from that point onwards.
Question 8.
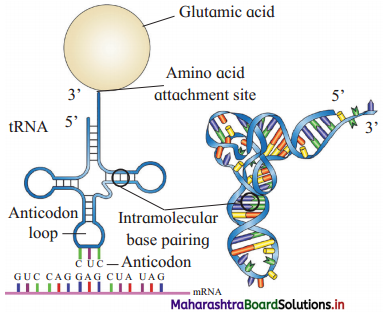
Transfer-RNA.
Answer:
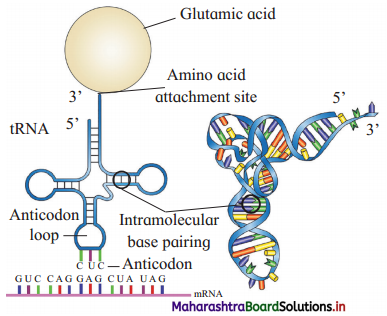
(1) As t-RNA can read the codon and also can bind with the amino acid, t-RNA is considered as an adapter molecule.
(2) Clover leaf structure (2 dimensional) of t-RNA:
- t-RNA has four arms – DHU arm (has amino acyl binding loop), middle arm (has anticodon loop), Tif/C arm (has ribosome binding loop) and variable arm.
- It has G nucleotide at 5’ end.
- Amino acid acceptor end (3’ end) having unpaired CCA bases (i.e. amino acid binding site).
(3) For every amino acid, there is specific t-RNA.
(4) Initiator t-RNA is specific for methionine.
(5) There are no t-RNAs for stop codons.
(6) In the actual structure, the t-RNA molecule looks like inverted L (3 dimensional : structure)
Question 9.
UTRs.
Answer:
- m-RNA has some additional sequences that are not translated. These sequences are referred as untranslated regions (UTR).
- The UTRs are present at both 5′-end (before start codon) and at 3′-end (after stop codon).
- They are required for efficient translation process.
Short Answer Questions
Question 1.
Why are Okazaki fragments formed on lagging strand only?
Answer:
- The lagging template is the template strand with free 5’ end.
- The replication always starts at C-3 end of template strand and proceeds towards C-5 end.
- Both the strands of the parental DNA are antiparallel and new strands are always formed in 5′ → 3′ direction, i.e. DNA polymerase synthesizes new strand in only one direction i.e. 5′ → 3′ direction.
- Hence, the lagging templates becomes available for replication only discontinuously in small patches.
- The new lagging strand develops discontinuously away from the replicating fork in the form of small Okazaki fragments.
- Hence, Okazaki fragments formed on lagging strand only.
Question 2.
Why t-RNA is called as adapter molecule?
Answer:
t-RNA can read the codon on m-RNA. It also can bind with the amino acid at 3’ end and transport it to m-RNA-ribosome complex during translation. It can bind with specific codon which is complementary to its anticodon. So t-RNA is considered as an adapter molecule.
Question 3.
Why DNA replication is called semiconservative replication?
Answer:
- In each of the two daughter DNA molecules thus formed, one strand is parental and the other one is newly synthesized.
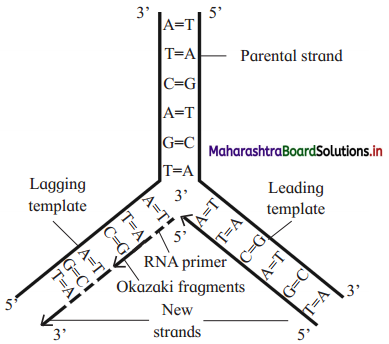
- 50% is contributed by mother DNA.
- Hence, DNA replication is described as semiconservative replication.
![]()
Question 4.
What are the functions of three types of RNAs in bacteria? Which enzyme is involved in transcription of DNA to form RNAs in bacteria? What is its function?
Answer:
- In bacteria, m-RNA provides the encoded message for protein synthesis; t-RNA brings specific amino acid to the site of translation; r-RNA plays role in providing binding site to m-RNA and t-RNA.
- There is single DNA dependent-RNA polymerase that catalyses transcription of all 3 types of RNA in bacteria.
- RNA polymerase binds to promoter and initiates transcription (initiation) and synthesizes RNA.
Question 5.
Explain why it was suggested that codon is a sequence of three consecutive nucleotides on m-RNA.
Answer:
- If each codon has only one nucleotide, then there will be 41 = 4 codons, which can encode for only four different types of amino acids.
- If each codon has two nucleotides, then there will be 4² = 16 codons, which can encode for only 16 different types of amino acids.
- If each codon has three nucleotides, then there will be 4³ = 64 codons, which are sufficient to specify 20 different types of amino acids.
Question 6.
Explain Genetic code is Non-overlapping.
Answer:
- Each single base is a part of only one codon.
- Adjacent codons do not overlap.
- If it had been overlapping type, with 6 bases, there would be 4 amino acid molecules in a chain.
- Experimental evidence favours non-overlapping nature of genetic code.
Question 7.
How degeneracy of the code is explained by Wobble hypothesis?
Answer:
- In 1966, Crick proposed Wobble hypothesis.
- According to this hypothesis, in codon- anticodon base pairing, the third base may not be complementary.
- The third base of codon is called wobble base and this position is called wobble position.
- This results in economy of t-RNA as anticodon of a t-RNA may bind with codon even when only first two bases are complementary.
- For example, GUU, GUC, GUA and GUG codons code for amino acid Valine.
- Degeneracy of genetic code means many codons can code for same amino acid.
- Thus, the degeneracy of genetic code gets explained by Wobble hypothesis.
Question 8.
a. What is meant by universal genetic code? Give example.
b. Why genetic code is called Non- ambiguous?
Answer:
a. Universal genetic code means that the specific codon codes for same amino acid in all living organisms, e.g. codon AUG always specifies amino acid methionine.
b. Genetic code is called non-ambiguous because, each codon specifies a particular amino acid.
Question 9.
Give examples of termination codons. Why are they known as termination codons?
Answer:
- UAA, UAG and UGA are known as termination codons.
- They do not code for any amino acid.
- They terminate or stop the process of elongation of polypeptide chain.
- Hence, they are known as termination codons.
Question 10.
How many amino acids are required for protein synthesis? From where are they obtained?
Answer:
- About 20 different types of amino acids are required for protein synthesis.
- They are available in the cytoplasm.
Question 11.
How DNA regulates protein synthesis?
Answer:
- DNA regulates protein synthesis by coding for the specific sequence of amino acids in a protein.
- This control is possible through transcription of m-RNA.
- Genetic code is specific for particular amino acid.
Question 12.
What is the role of ribosomes in protein synthesis?
Answer:
- Ribosomes serve as site for protein synthesis.
- A ribosome has one binding site for m-RNA and 3 binding sites for t-RNA. They are P-site (peptidylt-RNA-site), A-site (aminoacyl t-RNA-site) and E-site (exit site).
- In Eukaryotes, a groove which is present between two subunits of ribosomes, protects the polypeptide chain from the action of cellular enzymes and also protects m-RNA from the action of nucleases.
Question 13.
Give examples of coordinated regulation or expression, of several sets of genes.
Answer:
Examples of coordinated regulation or expression of several sets of gene are:
- If E.colt bacteria do not have lactose in the surrounding medium as a source of energy, then structural genes in Lac operon do not get transcribed and enzyme like β -galactosidase is not synthesized.
- The development and differentiation of embryo into an adult organism.
Question 14.
Explain with example what is meant by positive control of gene regulation.
Answer:
- A set of genes will be switched on when a substrate is to be metabolized.
- This phenomenon is called induction and small molecule responsible for this, is known as inducer.
- It is positive control of gene regulation.
- For example, lactose acts as an inducer in Lac operon.
Question 15.
If operator gene is deleted due to mutation, how will E.coli metabolise lactose?
Answer:
If operator gene is deleted due to mutation, lac operon cannot be regulated. It will get transcribed continuously and enzymes required for lactose metabolism will get synthesized continuously.
Question 16.
Explain in brief the process of initiation during protein synthesis.
Answer:
Initiation of synthesis of polypeptide chain takes place as follows:
- Small subunit of ribosome binds to the m-RNA at 5’ end.
- Start codon is positioned properly at P-site.
- Initiator t-RNA, (carrying methionine in eukaryotes or formyl methionine in prokaryotes) binds with initiation codon (AUG) of m-RNA by it’s anti-codon (UAC). Codon-anti-codon pairing involves formation of hydrogen bonds.
- The large subunit and smaller subunit of ribosome bind in the presence of Mg++.
- Thus, initiator charged t-RNA occupies the P-site and A-site is vacant for next charged t-RNA.
Question 17.
What are the application of genomics?
Answer:
Applications of genomics are as follows:
- Structural and functional genomics is used in the improvement of crop plant, human health and livestock.
- The knowledge and understanding acquired by genomics research can be applied in medicine, biotechnology and social sciences.
- It helps in the treatment of genetic disorders through gene therapy.
- It helps in the development of transgenic crops having desirable characters.
- Genetic markers have applications in forensic analysis.
- Genomics can lead to introduction of new gene in microbes to produce enzymes, therapeutic proteins and biofuels.
Question 18.
Why is HGP important?
Answer:
- HGP is associated with rapid development of Bioinformatics.
- Knowledge gained about the functions of genes and proteins has a major impact in the fields like Medicine. Biotechnology and the Life sciences.
- It has helped in identifying the genes that are associated with genetic characteristics.
- The genetic basis of many hereditary diseases can be understood.
- It has increased the understanding of gene structure and function in other species. As human beings have many of the genes same as those of flies, roundworms and mice, such studies will enhance understanding of human evolution.
Chart based / Table based Questions
Question 1.
Complete the following table:
| Organism | Diploid chromosome number |
| Mouse | ————- |
| Fruitfly | ———— |
| Roundworm | ————- |
| Yeast | ————– |
Answer:
| Organism | Diploid chromosome number |
| Mouse | 40 |
| Fruitfly | 8 |
| Roundworm | 12 |
| Yeast | 32 |
Diagram Based Questions
Question 1.
a. Identify A and B in the following diagram.
b. Name the scientist who conducted this experiment.
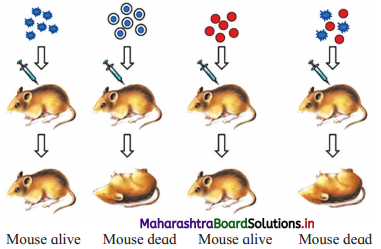
Answer:
a. A : Smooth strain (III-S)
B : Rough Strain and Heat-killed Smooth Strain
b. F. Griffith conducted the experiment shown in the diagram.
Question 2.
a. Identify A and B in the given diagram.
b. What is the conclusion of the given experiment?
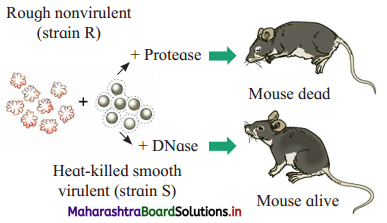
Answer:
a. A : Rough, nonvirulent R-strain B : Heat-killed virulent S-strain
b. When DNA of heat-killed S-strain bacteria is treated with DNase, mouse remains alive as transformation does not take place. This proves that DNA is the genetic material.
Question 3.
Identify A, B, C and name the scientists who carried out the experiment given in the diagram.
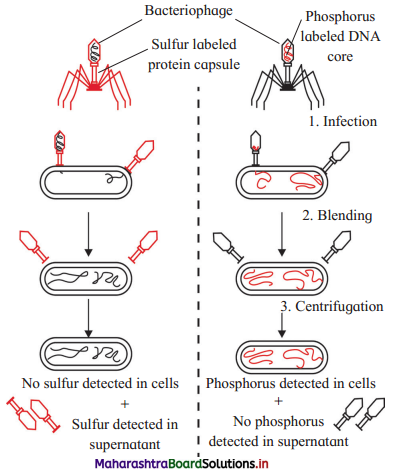
Answer:
Answer: A : Infection B : Blending C : Centrifugation Scientists who carried out experiment are Alfred Hershey and Martha Chase.
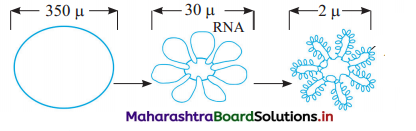
Question 4.
(1) Identify A, B and C.
(2) What are their sizes?
Answer:
(1) A : Circular, unfolded chromosome
b : Folded chromosome (40 to 50 loops)
c : Supercoiled, folded chromosome
(2) (A) 350 µ (B) 30 µ (C) 2 µ
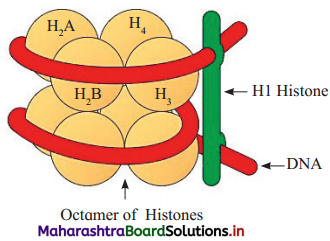
Question 5.
Draw a labelled diagram of Nucleosome with H1 histone.
Answer:
Question 6.
Identify A and B in the given diagram.
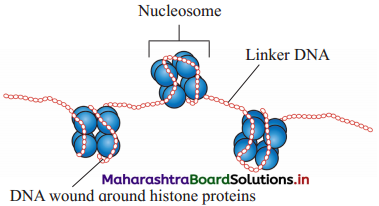
Answer:
A : Nucleosome
B : Linker DNA
Question 7.
(a) Identify A in the given diagram.
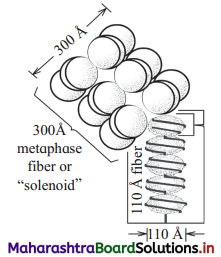
(b) What is the dimension denoted in ‘B’
Answer:
A : Solenoid B : 300 Å
Question 8.
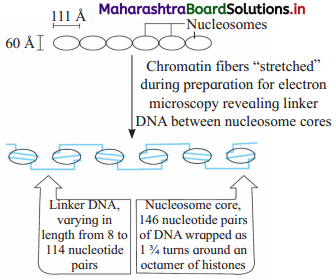
Sketch and label process of formation of beads on strings/fibres from chain of nucleosomes.
Answer:
Question 9.
a. Identify A. Which enzyme joined them?
b. Identify B. What is its function ?
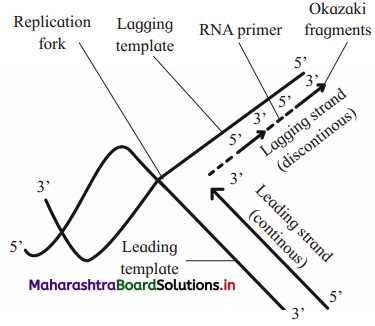
Answer:
a. A is Okazaki fragment. These fragments are joined by DNA ligase enzyme
b. B is RNA primer. It provides 3’ OH to which nucleotide gets attached by ester bond.
Question 10.
Draw a labelled diagram showing semi-conservative replication of DNA.
Answer:
![]()
Question 11.
Sketch and label, Meselson and Stahls experiment.
Answer:
Question 12.
Identify A, B and C in the following diagram.
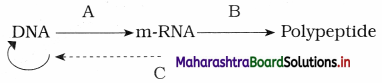
Answer:
A : Transcription
B : Translation and
C : Reverse Transcription
Question 13.
a. Draw a labelled diagram of transcription unit.
b. What is the sequence of m-RNA and coding strand if sequence of template strand of DNA is
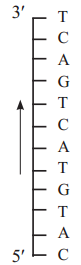
Answer:
a. 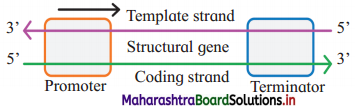
b. 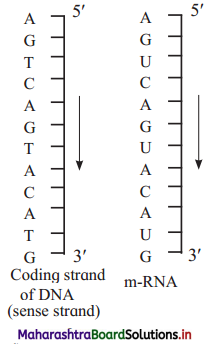
Question 14.
a. The following diagram shows processing of ………….
b. What is capping?
c. Identify A, B and C.
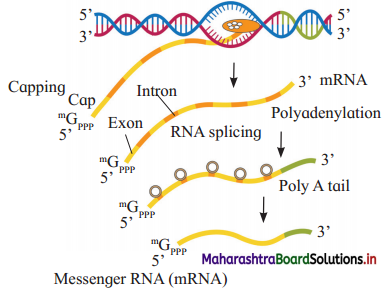
Answer:
a. hnRNA
b. Capping is addition of methylated guanosine tri-phosphate at 5′ end of hnRNA.
c. A : Exon and B : Intron C : m-RNA
Question 15.
Draw a labelled diagram of t-RNA carrying Glutamic acid.
Answer:
Question 16.
Observe the diagrams (a), (b) and (c)
- Which step of protein synthesis is shown in the following diagrams?
- During initiation, which subunit of ribosome binds with m-RNA?
- What are the three binding sites for t-RNA on ribosomes?
- On which site of ribosome second and subsequent t-RNA arrives?
- Which link is binding amino acids in diagram (b)?
- Which chain is being released from ribosome in diagram (c) ?
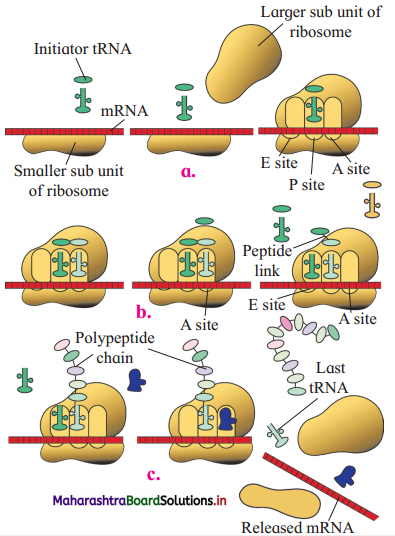
Answer:
- Translation
- 30S or 40S
- P site, A site and E site
- A site
- Peptide link
- Polypeptide chain
Question 17.
Draw a labelled diagram – Working of lac operon.
Answer:
Question 18.
What is the process shown in the following diagram? Mention all the steps given in the diagram in a proper sequence.
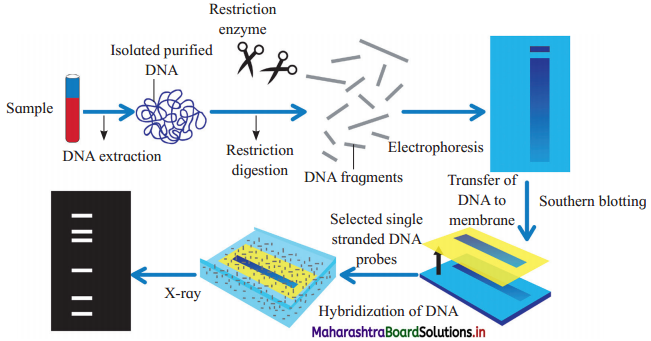
Answer:
The process shown in the above diagram is DNA fingerprinting. The sequence of steps in the process are:
- Isolation of DNA
- Restriction digestion
- Gel electrophoresis
- Southern blotting
- Selection of DNA probe
- Hybridization
- Photography,
Long Answer Questions
Question 1.
Describe Griffith’s transformation experiment.
OR
In the light of Griffith’s experiment, explain the action of two strains of streptococcus pneumoniae and give his conclusion.
Answer:
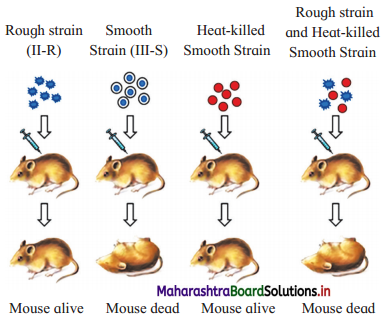
(1) In 1928, Frederick Griffith, carried out experiments with bacterium Streptococcus pneumoniae (which causes pneumonia in humans and other mammals).
(2) Griffith used two strains of Streptococcus pneumonia:
- S-type (Virulent, smooth, pathogenic and encapsulated).
- R-type (Non-virulent, rough, non- pathogenic and non-capsulated).
(3) Experiments carried out by E Griffith:
- Mice were injected with R-strain bacteria and they survived (no pneumonia).
- Mice injected with S-strain bacteria developed pneumonia and died.
- When heat-killed S-strain bacteria were injected in mice, the mice survived.
- On injecting a mixture of heat-killed S-bacteria and live R bacteria, the mice died.
(4) Griffith obtained live S-strain bacteria from the blood of the dead mice.
(5) He concluded that the live R-strain bacteria must have picked up something (which he called transforming principle) from the heat killed S bacterium, and got changed into S-type. Transforming principle allowed R-type to synthesize capsule and it became virulent.
(6) Thus, F. Griffith first demonstrated genetic transformations.
Question 2.
Describe Avery, McCarty and MacLeod’s experiments.
Answer:
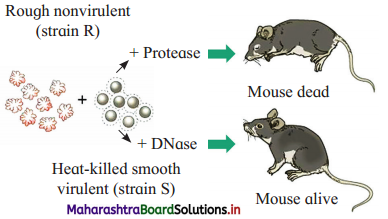
- In 1944, Oswald T. Avery, Colin M. MacLeod and Maclyn McCarty proved that the DNA is a genetic material (transforming principle).
- They purified DNA, RNA, proteins (enzymes) and other materials from heat killed S-strain cells and mixed them with cells of R-strain bacteria separately to confirm which one could transform living R cells into S cells.
- Only DNA was able to transform harmless R-strain into virulent S-strain.
- They also demonstrated that proteases, RNases did not affect transformation. Thus it was proved that the transforming substance was neither a protein nor-RNA.
- When DNA was digested with DNase, there was no transformation.
- These experiments proved that the transformation of Live R-strain bacteria into S-strain type was because of DNA of bacteria of S-strain.
- Thus, they proved that the DNA is transforming principle.
Question 3.
Explain how Hershey – Chase experimentally proved that DNA is the genetic material.
Answer:
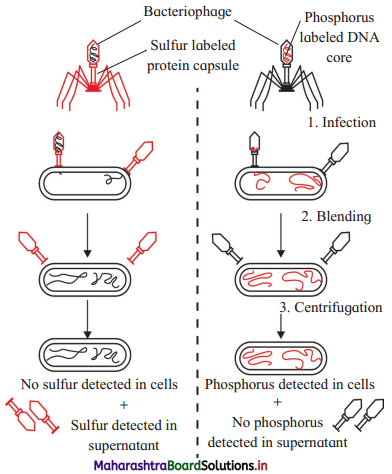
(1) Hershey and Chase worked with bacteriophages (viruses that infect bacteria and which are composed of DNA and protein coat).
(2) They cultured E. coli bacteria in medium containing radioactive phosphorus 32p. By infecting these bacteria with bacteriophages, Hershey and Chase could develop bacteriophages having DNA labelled with 32p. as DNA contains phosphorus and proteins do not.
(3) They also cultured E. coli bacteria in medium containing radioactive sulphur 35s. By infecting these bacteria with bacteriophages, they developed
bacteriophages whose protein coat was labelled with 35s, as proteins contain sulphur and DNA does not.
[Note : Viruses cannot be cultivated in medium.]
(4) Experiment involved three steps.
- Infection : Both the types of radioactive phages were allowed to infect E.coli bacteria grown on the medium containing normal ‘P‘ and ‘S’.
- Blending : Then bacterial cultures were agitated in blender to break contact between bacteria and parts of viruses that did not enter bacterial cells.
- Centrifugation : It was done to separate bacterial cells as a pellet. Parts of viruses which did not enter bacteria remained in the suspension.
(5) Observation:
- Radioactive ‘S’ remained in suspension.
- Only radioactive ‘P’ was found inside the bacterial cell in the pellet.
(6) Thus it was proved that DNA is the genetic material which enters bacterial cell and not protein.
![]()
Question 4.
Explain the formation of beads on string, solenoid fibre, chromatin fibre and chromosome.
Answer:
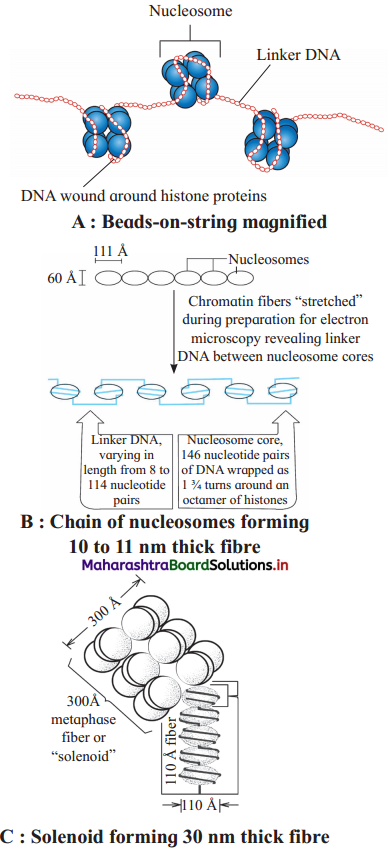
- Beads on string (11 nm in diameter) : Under an electron microscope, nucleosomes in thread like chromatin look like ’beads-on- string’.
- Solenoid fibre (30 nm in diameter) : Six such nucleosomes get coiled and then form solenoid that looks like coiled telephone wire.
- Chromatin fibre (200 nm in diameter) : Further supercoiling tends to form a looped structure called chromatin fibre.
- Chromosome (1400 nm in diameter) : Chromatin fibre further coils and condenses at metaphase stage to form the chromosomes. Each chromatid is 700 nm in diameter.
- Non-Histone Chromosomal Proteins (NHC) are the additional sets of proteins that contribute to the packaging of chromatin at a higher level.
Question 5.
Explain the components of a transcription unit with the help of a diagram?
Answer:
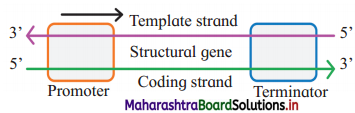
Transcription unit (Each transcribed segment of DNA) consists of the promoter, the structural gene and the terminator.
(1) The promoter:
- The promoter is located towards 5′ end of structural gene, i.e. upstream.
- It is a DNA sequence that provides binding site for enzyme RNA polymerase.
- In prokaryotes, sigma factor sub unit of the enzyme recognizes the promoter.
(2) Structural genes:
- Template strand (Antisense strand) : DNA strand having 3’→ 5’ polarity acts as template strand as DNA dependent- RNA polymerase catalyses polymerization in 5’ → 3’ direction.
- Sense strand : The other strand of DNA having 5’ → 3’ polarity is complementary to template strand. It is called as sense strand. The sequence of bases in this strand, is same as in RNA (where Thymine is replaced by Uracil). It is the actual coding strand.
(3) The terminator:
- The terminator is located at 3’ end of coding strand, i.e. downstream.
- It defines the end of the transcription process.
Question 6.
What have we learnt from the Human Genome Project?
Answer:
We have learnt following salient features of human genome from the Human Genome Project.
- The human genome contains 3164.7 million nucleotide bases.
- The average gene consists of 3000 bases.
- Largest known human gene is dystrophin at 2.4 million bases.
- Total number of genes is estimated to be 3000.
- 99.9% nucleotide bases are exactly the same in all people.
- The function of about 50% of the discovered genes are unknown.
- Less than 2% of the genome codes for proteins.
- Repeated sequences make up a very large portion of the human genome. They can shed light on chromosome structure, dynamics and evolution.
- Chromosome 1 has most genes (2968) and the Y has the fewest (231).
- Single nucleotide polymorphism have been identified at about 1.4 million locations. It is useful in finding chromosomal locations for disease-associated sequences and tracing human history.
Question 7.
Describe the steps involved in DNA fingerprinting ?
Answer:
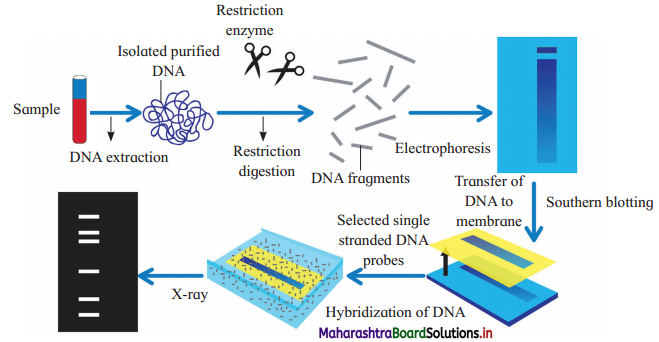
Steps involved in DNA fingerprinting are as follows:
1. Isolation of DNA : The DNA can be isolated even from the small amount of tissue like blood, hair roots, skin, etc.
2. Restriction digestion:
- The isolated DNA is treated with restriction enzymes which cut the DNA at specific sites to form small fragments of variable lengths.
- Variations in the lengths of restriction fragments are known as Restriction Fragment Length Polymorphism (RFLP).
3. Gel electrophoresis:
- The DNA samples are loaded on agarose gel and electrophoresis is carried out.
- Negatively charged DNA fragments move to the positive pole.
- Separation of fragments depends on their length and it results in formation of bands.
- dsDNA is then denatured into ssDNA by alkali treatment.
4. Southern blotting : The separated DNA fragments are transferred to a nylon membrane or a nitrocellulose membrane.
![]()
5. Selection of DNA probe:
- DNA Probe is a known sequence of single- stranded DNA.
- It is obtained from organisms or prepared by cDNA preparation method.
- The DNA probe is labelled with radioactive isotopes.
6. Hybridization:
- In this process, probe is added to the nitrocellulose membrane containing DNA fragments.
- The single-stranded DNA probe pairs with the complementary base sequence of the DNA strand.
- As a result DNA-DNA hybrids are formed on the nitrocellulose membrane. Unbound single-stranded DNA probe fragments are washed off.
7. Photography : The nitrocellulose membrane is then kept in contact with X-ray film. DNA bands, due to radioactive probe, give photographic image on X-ray film. This is autoradiography.